Advancements in Genome Sequencing Over The Last 2 Decades
Genomics is an interdisciplinary field of biology that involves sequencing and study of the human genome. It gives the complete information of the particular organisms DNA and genes present in it. The studies in several scientific disciplines depend on the use of DNA analysis to understand theories and assumptions at a genetic and genomic level. With that very strong reliance on these techniques, it is no surprise that the science of genetic analysis or genomics is quickly evolving and developing technologies that are extremely efficient and useful. Even though most of us share approximately 99.5 percent of our DNA but still there are millions of variations between the genome of two different individuals. Genome sequencing can determine the vulnerability, ancestry, characteristics, and response to drugs or therapies and helps create novel selective therapies, also known as precision medicines. The genetic basis of about 60 disorders was known to researchers before Human Genome Project. But today, they have complete genetic information of around 5000 disorders.
The progressive evolution of genome sequencing technologies with the reduction in cost and time has been observed over the last two decades. The first commercialised technique for DNA sequencing was the Sanger method. It was developed in 1977 by Laureate Frederick Sanger for which he was awarded Nobel Prize in 1980. The Human genome project, which was launched in the year 1990 to sequence the entire genome of the human cell was completed in 2003 and is considered a significant achievement in the field of science. But it took around 13 years to sequence the human genome using this technique and the total cost of the project was approximately 2.7 billion US dollars. Completed in 2012, the “1000 Genomes Project” sequenced 1092 genomes so that we can somehow understand more about human genome variation and diversity.
After the phenomenal success of the Human Genome Project, there has been significant development of Second-Generation Sequencing techniques which is also known as Next-Generation Sequencing (NGS). New technologies have been introduced to speed up the sequencing process, enabling parallel sequencing of numerous DNA fragments through different chemical reactions and optical detection methods. These techniques are capable of providing significantly higher sequencing throughout and thereby give a lower price per sequenced base. Billions of DNA nucleotides can be sequenced at a single point of time and it also limits the requirement for the part of cloning strategies that were used in Sanger sequencing. Some of the next-generation sequencing techniques include Illumina Genome Analyser, Applied BioSystems SOLiD, Roche 454 pyro-sequencing, Complete Genomics, Pacific Biosciences, Helios, and IonTorrent. The first, commercially successful second-generation sequencing technique was Roche 454 pyro-sequencing which was developed in 2005 by 454 Life Science. The estimated cost of sequencing by this method is $10 per million bases. One of the major drawbacks of NGS is that it is unable to read the entire DNA sequence of the genome, they are restricted to small DNA fragments sequencing and produce millions of reads.
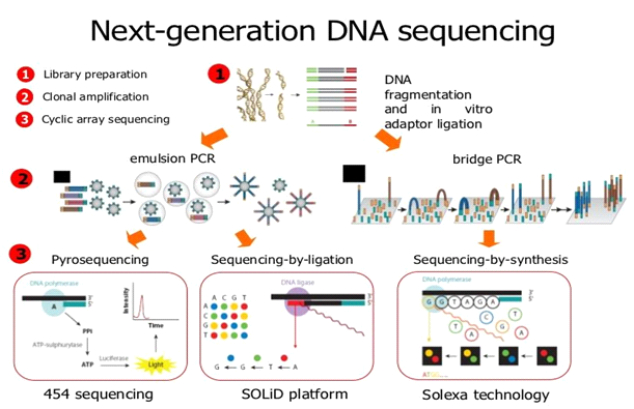
Currently, third-generation sequencing is an emerging technology for single-molecule sequencing. To reduce the price of sequencing and to automate preparatory procedures and methods of sequencing, these techniques have been developed. Third-generation sequencing techniques include Single-molecule real-time sequencing, Helicos sequencing, etc. One of the most widely used third-generation sequencing techniques is nanopore sequencing. It was made commercially available in 2015. It is a type of long-read sequencing system that can explicitly read long nucleotide sequences. It differs from the second-generation sequencing technique which cuts the DNA into small strands, sequences them, and then uses algorithms to combine the fragments into a complete sequence. In this, a molecule is forced to pass through a very small cavity and the detectors detect the bases as it passes along. This technique greatly reduces the cost of sequencing and the results can then be generated in about a week.
We used to think that our fate was in our stars. Now we know that, in large measure, our fate is in our genes.
– Francis Crick
The future of genome analysis or sequencing will continue to offer us exciting advancement and novel results in a wide variety of research fields. The continuous upward trajectory of sequencing technology advancement allows diagnostic methods to be enhanced. These days, DNA sequencing techniques are being used to diagnose complex and mysterious neuro-developmental disorders. They are also used to sequence tumors so that new personalised therapies can be developed. These techniques got so advanced that pregnant women can now avoid invasive techniques like amniocenteses to investigate chromosomal aberrations in the fetus and can get the results by a simple blood test. The availability of cheap, high-performance sequencing techniques will continue to increase the diversity of genomic research and applications. In the future, the goal of researchers will be to increase efficiency, accuracy and to reduce the processing time. The data obtained by sequencing are used by scientists for the development of new diagnostics and prognostic techniques.


 Kindly fill the form below to download our Liquid Biopsy Portfolio brochure :
Kindly fill the form below to download our Liquid Biopsy Portfolio brochure :